import numpy as np
import matplotlib.pyplot as plt
def ackley(x, a=20, b=0.2, c=2*np.pi):
d = len(x)
return -a * np.exp(-b * np.sqrt(np.sum(np.square(x))/d)) - \
np.exp(np.sum(np.cos(c * x))/d) + a + np.exp(1)Week 8 lab
Set up
Test different approaches to generating the objective function surface
# Method 1: list comprehension (2D)
def method1(n):
x = np.linspace(-30, 30, n) # n points between -30 and 30
y = np.linspace(-30, 30, n) # n points between -30 and 30
return (x, y, np.array([[ackley(np.array([i, j])) for i in x] for j in x])) # 2D array using a list comprehension
%timeit method1(501)3.75 s ± 75.2 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)# Method 2: pre-allocated array (2D)
def method2(n):
x = np.linspace(-30, 30, n) # n points between -30 and 30
y = np.linspace(-30, 30, n) # n points between -30 and 30
z = np.zeros((n, n)) # pre-allocate the 2D array
for i in range(n):
for j in range(n):
z[i, j] = ackley(np.array([x[i], y[j]]))
return (x, y, z)
%timeit method2(501)3.79 s ± 47.6 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)# Method 3: meshgrid with list comprehension (1D)
def method3(n):
x = np.linspace(-30, 30, n)
y = np.linspace(-30, 30, n)
(xx, yy) = np.meshgrid(x, y) # 2D arrays of x and y values separately
xy = np.column_stack((xx.flatten(), yy.flatten())) # 2D array of all combinations of x and y; first column is x and second column is y
return (x, y, np.array([ackley(xyi) for xyi in xy]).reshape(xx.shape))
%timeit method3(501)3.79 s ± 249 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)Summary
There is no discernable difference between the different methods - use whichever comes most naturally!
Plotting
# plot contour lines
(x, y, z) = method1(1001)
plt.figure()
plt.contour(x, x, z, levels=np.linspace(0, 20, 21), colors='black')
plt.show()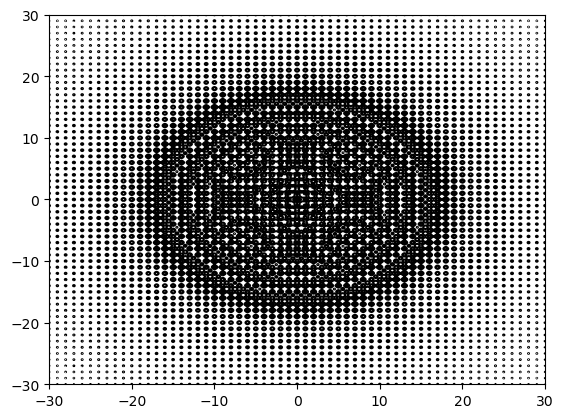
Julia
Corresponding Julia code (same as method 1 in Python) is given by
function method1(n)
x = range(-30, 30, length=n)
y = range(-30, 30, length=n)
return [ackley([xi, yi]) for xi in x, yi in y]
endwhich when timed gives
julia> using BenchmarkTools
julia> @btime method1(501)
34.832 ms (753005 allocations: 59.36 MiB)which is 108 times faster. A small modification to use statically-sized arrays gives a further speed up
using StaticArrays
function method2(n)
x = range(-30, 30, length=n)
y = range(-30, 30, length=n)
return [ackley(@SVector[xi, yi]) for xi in x, yi in y]
endwhich when timed gives
julia> @btime method2(501)
8.870 ms (2 allocations: 1.92 MiB)which is now 427 times faster than Python. Summary: Python is slow…